Table of Content
FFPE Transcriptomics Offers Valuable Insights into Diseases
3′ mRNA-Seq vs Whole Transcriptome RNA-Seq
3′ mRNA-Seq is Ideal for Expression Profiling
Whole Transcriptome RNA-Seq for Isoform and Biomarker Analysis
Which Method to Choose for RNA-Seq from FFPE Samples?
3′ mRNA-Seq with QuantSeq: Showcases
Whole Transcriptome RNA-Seq with CORALL: Showcases
FFPE Transcriptomics Offers Valuable Insights into Diseases
Transcriptome profiling gives insights into gene expression during disease progression and all possible alterations to the transcripts present in healthy versus diseased tissues. Having information on transcriptional profiles of tissues and cells is becoming increasingly important in clinical decision-making and academic research. Analyzing changes in gene expression allows us to understand the disease-induced changes. More complex whole transcriptome analyses may follow if the underlying mechanisms are of interest.
Various diseases have been associated with changes in alternative polyadenylation site usage, splicing malfunctions, and malignant transcript variants with nucleotide exchanges, insertions or deletions, as well as fusion transcripts (Bessa et al., 2020; Dorney et al., 2023). These variations are not always associated with coding transcripts. They can also affect the non-coding transcriptome, including long non-coding RNAs (lncRNAs, French and Edwards, 2020). Many lncRNAs are differentially expressed in disease conditions implying their fundamental role in various biological processes. This is why they became important biomarkers for pathological states.
Many different patient sample types are suitable for transcriptome profiling studies including various biofluids, such as whole blood, plasma, cerebrospinal fluid, urine, and all possible solid tissues (e.g., biopsies from brain, skin, liver, kidneys, etc.). When it comes to preserving RNA in tissue biopsies, fixing in formalin and embedding in paraffin (FFPE samples) and flash freezing (fresh frozen, FF samples) are the most common tissue preservation techniques. In our recent blog, we compared fresh frozen and FFPE samples for next-generation sequencing studies.
While RNA from frozen samples is of higher quality than RNA extracted from FFPE samples, FFPE samples have other advantages that make them more suitable despite being more challenging to work with. Estimates are that around 50 to 80 million of the FFPE samples stored globally may be suitable for NGS analysis, thus enabling retrospective studies due to their well described nature. Sample information includes primary diagnosis, therapeutic regimen, drug response, and recurrence — so these collections of FFPE samples have enormous potential for understanding and treating various human diseases. Read more on the applications of FFPE samples in modern research in one of our recent blogs.

RNA-Seq is a state-of-the-art approach for transcriptome profiling of FFPE samples. The main bottleneck of performing RNA-Seq from FFPE samples is obtaining RNA of sufficient quality for further processing owing to fragmentation and cross-linking to proteins introduced during the fixation process. Luckily, nowadays, there are many workarounds, optimized protocols, products specifically developed for extracting RNA from FFPE samples, and RNA-Seq library preparation of FFPE-RNA. In addition, knowledge is growing on how to approach RNA-Seq data analysis for libraries stemming from FFPE material.
Bulk RNA-Seq approaches to studying the transcriptome are 3’ mRNA sequencing and whole transcriptome sequencing (sometimes referred to as Total RNA-Seq). 3′ mRNA-Seq technologies are primarily designed for gene expression profiling. On the other hand, whole transcriptome RNA-Seq, as the name suggests, is designed for the analysis of the entire transcriptome. Both methods are commonly used for transcriptomics from archived FFPE samples; choosing the right approach will depend on the area of interest and research questions.
In this blog, we will explain the basics of 3’ mRNA-Seq and whole transcriptome sequencing, when to use which of these RNA-Seq approaches, and end with showcasing several studies using Lexogen products for 3’ mRNA-Seq and whole transcriptome RNA-Seq on FFPE samples.
3' mRNA-Seq vs Whole Transcriptome RNA-Seq on FFPE samples
FFPE samples are most commonly (but not exclusively) used in cancer research and oncology. Cancer researchers often need information on differential gene expression in tissues collected through biopsies from cancer patients. Following biopsies, the tissue is usually fixed and embedded in paraffin to stabilize it for decades. These FFPE blocks can be archived in biobanks which are accessible to cancer researchers. FFPE samples are often used due to their abundance, availability, and association with patient outcomes. However, these samples are extremely precious and harbor highly compromised, low-quality, degraded RNA. Therefore, working with them requires a lot of experience. In addition, appropriate RNA-Seq library preparation kits are needed, with exceptional sensitivity and the ability to generate libraries from degraded RNA samples. Both whole transcriptome sequencing (WTS) and 3′ mRNA-Seq can be used for differential expression analysis of FFPE samples.
3′ mRNA-Seq is Ideal for Expression Profiling
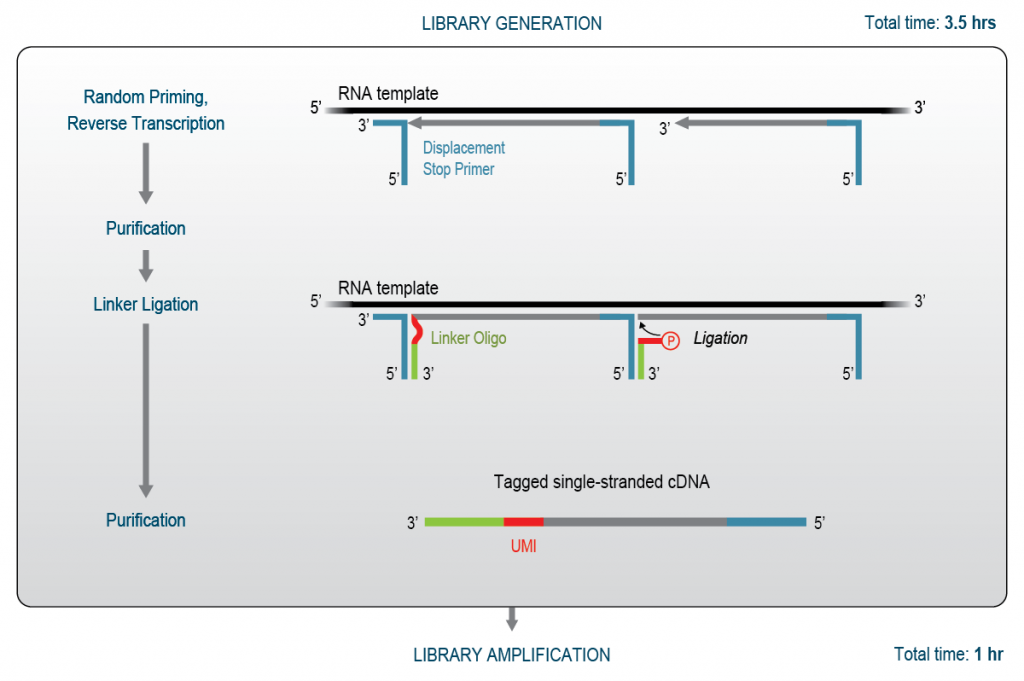
3′ mRNA-Seq is ideal for differential gene expression. This method focuses sequencing reads on the 3’ ends of all polyadenylated transcripts. Typically, oligo(dT) primers are used to initiate reverse transcription (also referred to as first strand cDNA synthesis). Afterwards, the RNA template is removed to enable a random second strand synthesis primer to bind to the cDNA and generate a double-stranded cDNA library. Figure 1 shows the workflow used for QuantSeq – the first 3′ mRNA-Seq kit introduced in 2014. Variations of this workflow, e.g., using template switching and tagmentation at the 3′ end are also possible. Several methods, including QuantSeq FFPE, additionally contain unique molecular identifiers (UMIs) for more precise quantification. Finally, the resulting double-stranded cDNA is amplified, whereby indices for sample identification are added and adapters for sequencing are introduced. As 3′ mRNA-Seq starts directly from total RNA by oligo(dT) priming, prior poly(A) enrichment or rRNA depletion are not required. This does not only efficiently shorten the workflow time, but also significantly reduces overall consumable and experiment costs. In addition, sequencing reads are concentrated at the 3′ end, reducing sequencing depth requirements and costs for data analysis and data storage.
Figure 1 | QuantSeq 3′ mRNA-Seq Library Generation Workflow. Oligo(dT) priming initiates cDNA generation from the 3′ end of polyadenylated mRNAs. Following removal of the RNA template, random-primed second strand synthesis generates double-stranded 3′ cDNA library molecules with partial sequencing adapters.
Whole Transcriptome RNA-Seq for Isoform and Biomarker Analysis
Whole transcriptome RNA-Seq is the most commonly used method to generate sequencing libraries. Here, reads are distributed across the complete transcript body. These methods rely on random primers to initiate cDNA synthesis and thus require enrichment of the RNA of interest prior to library preparation. Therefore, ribosomal RNA needs to be removed as random primers can bind rRNA and generate inserts from undesired, abundant RNAs cluttering the sequencing data. Figure 2 illustrates the library preparation workflow for CORALL RNA-Seq, a WTS library prep.
Figure 2 | CORALL RNA-Seq Library Generation Workflow. Random displacement stop primers (DSP) containing partial adapter sequences initiate reverse transcription and cDNA synthesis. Each reverse transcribed fragment is stopped at the downstream DSP. An efficient linker ligation step introduces a second partial adapter sequence and a UMI, resulting in a single-stranded cDNA library that is amplified to conclude NGS library preparation.
CORALL uses Lexogen’s proprietary displacement stop technology to generate NGS library inserts, without any RNA fragmentation steps (Fig. 2). Random displacement stop primers (DSPs) initiate cDNA synthesis by reverse transcriptase which is efficiently stopped when encountering a downstream DSP. The second step, linker oligo ligation, introduces UMIs. Both, reverse transcription and linker oligo ligation incorporate partial sequencing adapters and the resulting single-stranded cDNA library is then amplified. As for 3′ mRNA-Seq library generation, the final amplification step completes the adapter sequences for NGS and introduces indices to discriminate the samples during data analysis. Paired with RiboCop for enzyme-free rRNA depletion, the workflow is ideal for any FFPE application requiring coverage uniformity, including coverage analysis, alternative splicing or fusion gene detection and analysis of non-coding RNAs (e.g., lncRNA biomarkers).
Which Method to Choose for RNA-Seq from FFPE Samples?
After introducing the principles for both methodologies, the question remains which method to choose for RNA sequencing from FFPE samples. This choice depends on the experimental aim, i.e., if quantitative or qualitative data is required, on the RNA molecules of interest (e.g., mRNA. lncRNA, etc.), and economical factors around the project. For example, budget, sample availability, and data storage can also be important aspects to consider when deciding for one approach over another. Before diving into examples how the two methods have been used, we will give a short overview of how they compare to each other.
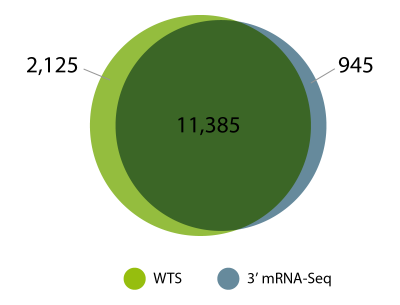
In our experiments, we consistently see a high degree of overlap in gene detection for FFPE samples processed with 3′ mRNA-Seq (using QuantSeq FFPE) and fragmentation-free whole transcriptome RNA-Seq (using CORALL FFPE, Fig. 3). For the experiment shown in Figures 3 and 4, the same input RNA from human kidney tumor FFPE material was processed with both methods. Sequencing data was then compared to assess which genes are detected by 3′ mRNA-Seq and WTS and how large the overlap of gene detection is. The results demonstrate that 3′ mRNA-Seq and whole transcriptome RNA-Seq are equally effective and suitable for looking at gene expression from archived FFPE samples. Similar observations were also previously reported by independent benchmarking studies comparing 3′ mRNA-Seq, targeted RNA sequencing and different whole transcriptome library preps (Turnbull et al., 2020).
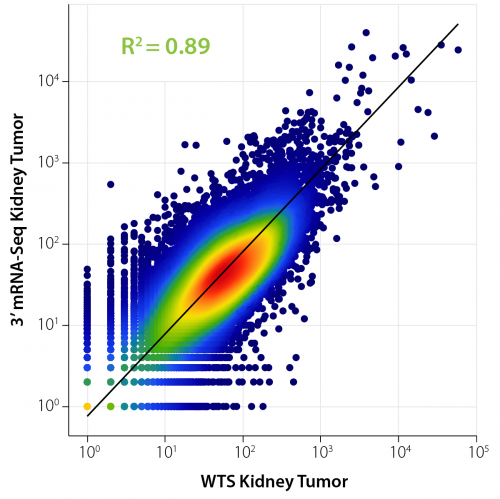
Correlation analyses are routinely used to assess differences and similarities between conditions. Figure 4 illustrates the correlation of expressed genes from the same sample (human kidney tumor FFPE RNA) prepared with two different library preparation methods: WTS and 3′ mRNA-Seq. A high correlation and thus high similarity of gene expression is indicated by the R² value of 0.89. The differences can partially be attributed to the use of rRNA depletion for the WTS prep which allows to assess also non-polyadenylated and non-coding transcripts which are not covered in 3′ mRNA-Seq libraries (see below).
Regarding sequencing depth, 3′ mRNA-Seq libraries can be sequenced at lower depths than WTS libraries, making data analysis particularly efficient and fast as there is a lower volume of data generated and no need to identify and normalize isoforms. Read counting provides accurate information on expression profiles with less computational resources and more rapid data turnaround, which is especially beneficial when working with a large number of samples. To resolve transcriptome isoforms, whole transcriptome RNA-Seq methods require deeper sequencing than 3′ mRNA-Seq methods, where reads are generated randomly, spanning all expressed isoforms of a given gene. As a result, data analysis of whole transcriptome libraries is more complex. More computational resources are required to assign the reads to the originating isoforms. Still, WTS provides a higher level of transcriptome information and is particularly useful to assess splicing events, fusions and to interrogate biomarkers for disease formation and progression.
Check our recent poster for an overview how 3′ mRNA-Seq and whole transcriptome sequencing can be integrated to identify hallmarks of kidney tumors from FFPE samples.
3' mRNA-Seq on FFPE Samples with QuantSeq: Showcases from Cancer Research
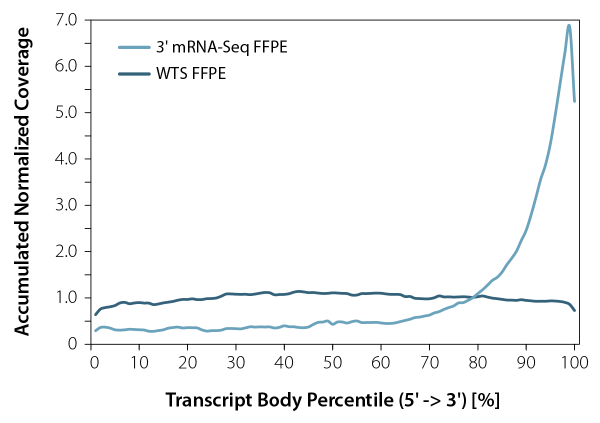
3′ mRNA-Seq library preparation kits, such as our QuantSeq, enable accurate gene expression quantification independently of the RNA quality. Standard mRNA-Seq protocols, which aim to cover the whole transcript, can lead to a significant 3’ bias when used on degraded RNA input, making them unsuitable for processing RNA from FFPE samples. This limitation underscores the necessity of using 3′ mRNA-Seq methods which are the only viable and cost-efficient tool to globally interrogate mRNA expression from low-quality samples, especially when high numbers of samples need to be processed and analyzed.
To date, QuantSeq has been successfully used for gene expression studies from FFPE material in hundreds of publications. We looked into our collection of publications and webinars featuring QuantSeq and prepared a small selection as a showcase. Find out how QuantSeq was used on FFPE samples for various applications in cancer research.
Publications using QuantSeq for 3′ RNA-Seq of FFPE Samples
Abstract
Overtreatment with cisplatin-based chemotherapy is a major issue in the management of muscle-invasive bladder cancer (MIBC), and currently none of the reported biomarkers for predicting response have been implemented in the clinic. Here we perform a comprehensive multi-omics analysis (genomics, transcriptomics, epigenomics and proteomics) of 300 MIBC patients treated with chemotherapy (neoadjuvant or first-line) to identify molecular changes associated with treatment response. DNA-based associations with response converge on genomic instability driven by a high number of chromosomal alterations, indels, signature 5 mutations and/or BRCA2 mutations. Expression data identifies the basal/squamous gene expression subtype to be associated with poor response. Immune cell infiltration and high PD-1 protein expression are associated with treatment response. Through integration of genomic and transcriptomic data, we demonstrate patient stratification to groups of low and high likelihood of cisplatin-based response. This could pave the way for future patient selection following validation in prospective clinical trials.
Read more to find out how QuantSeq was used to unravel chemotherapy response in muscle invasive bladder cancer.
Abstract
Uterine leiomyomas, or fibroids, are the most common tumors in women of reproductive age. Uterine leiomyomas can be classified into at least three main molecular subtypes according to mutations affecting MED12, HMGA2, or FH. FH-deficient leiomyomas are characterized by activation of the NRF2 pathway, including upregulation of the NRF2 target gene AKR1B10. Here, we have identified a novel leiomyoma subtype showing AKR1B10 expression but no alterations in FH or other known driver genes. Whole-exome and whole-genome sequencing revealed biallelic mutations in key genes involved in neddylation of the Cullin 3-RING E3 ligase, including UBE2M, NEDD8, CUL3, and NAE1. 3′RNA sequencing confirmed a distinct molecular subtype with activation of the NRF2 pathway. Most tumors displayed cellular histopathology, perivascular hypercellularity, and characteristics typically seen in FH-deficient leiomyomas. These results suggest a novel leiomyoma subtype that is characterized by distinct morphological features, genetic alterations disrupting neddylation of the Cullin 3-RING E3 ligase, and oncogenic NRF2 activation. They also present defective neddylation as a novel mechanism leading to aberrant NRF2 signaling. Molecular characterization of uterine leiomyomas provides novel opportunities for targeted treatment options.
Read how Quantseq was employed for the molecular characterization of uterine leiomyomas.
Abstract
Background
Preclinical research suggests that the efficacy of immune checkpoint inhibitors in breast cancer can be enhanced by combining them with antiangiogenics, particularly in a sequential fashion. We sought to explore the efficacy and biomarkers of combining the anti-PD-L1 durvalumab plus the antiangiogenic bevacizumab after bevacizumab monotherapy for advanced HER2-negative breast cancer.
Methods
Patients had advanced HER2-negative disease that progressed while receiving single-agent bevacizumab maintenance as a part of a previous chemotherapy plus bevacizumab regimen. Treatment consisted of bi-weekly durvalumab plus bevacizumab (10 mg/kg each i.v.). Peripheral-blood mononuclear cells (PBMCs) were obtained before the first durvalumab dose and every 4 weeks and immunophenotyped by flow-cytometry. A fresh pre-durvalumab tumor biopsy was obtained; gene-expression studies and immunohistochemical staining to assess vascular normalization and characterize the immune infiltrate were conducted. Patients were classified as “non-progressors” if they had clinical benefit (SD/PR/CR) at 4 months. The co-primary endpoints were the changes in the percentage T cell subpopulations in PBMCs in progressors versus non-progressors, and PFS/OS time.
Results
Twenty-six patients were accrued. Median PFS and OS were 3.5 and 11 months; a trend for a longer OS was detected for the hormone-positive subset (19.8 versus 7.4 months in triple-negatives; P = 0.11). Clinical benefit rate at 2 and 4 months was 60% and 44%, respectively, without significant differences between hormone-positive and triple-negative (P = 0.73). Non-progressors’ tumors displayed vascular normalization features as a result of previous bevacizumab, compared with generally abnormal patterns observed in progressors. Non-progressors also showed increased T-effector and T-memory signatures and decreased TREG signatures in gene expression studies in baseline—post-bevacizumab—tumors compared with progressors. Notably, analysis of PBMC populations before durvalumab treatment was concordant with the findings in tumor samples and showed a decreased percentage of circulating TREGs in non-progressors.
Conclusions
This study reporting on sequential bevacizumab+durvalumab in breast cancer showed encouraging activity in a heavily pre-treated cohort. The correlative studies agree with the preclinical rationale supporting an immunopriming effect exerted by antiangiogenic treatment, probably by reducing TREGs cells both systemically and in tumor tissue. The magnitude of this benefit should be addressed in a randomized setting.
Read on to find out how QuantSeq was used for gene expression studies in tumor samples for HER2-negative advanced breast cancer.
Abstract
The prostate cancer (PCa) field lacks clinically relevant, syngeneic mouse models which retain the tumour microenvironment observed in PCa patients. This study establishes a cell line from prostate tumour tissue derived from the Pten−/−/trp53−/− mouse, termed DVL3 which when subcutaneously implanted in immunocompetent C57BL/6 mice, forms tumours with distinct glandular morphology, strong cytokeratin 8 and androgen receptor expression, recapitulating high-risk localised human PCa. Compared to the commonly used TRAMP C1 model, generated with SV40 large T-antigen, DVL3 tumours are immunologically cold, with a lower proportion of CD8+ T-cells, and high proportion of immunosuppressive myeloid derived suppressor cells (MDSCs), thus resembling high-risk PCa. Furthermore, DVL3 tumours are responsive to fractionated RT, a standard treatment for localised and metastatic PCa, compared to the TRAMP C1 model. RNA-sequencing of irradiated DVL3 tumours identified upregulation of type-1 interferon and STING pathways, as well as transcripts associated with MDSCs. Upregulation of STING expression in tumour epithelium and the recruitment of MDSCs following irradiation was confirmed by immunohistochemistry. The DVL3 syngeneic model represents substantial progress in preclinical PCa modelling, displaying pathological, micro-environmental and treatment responses observed in molecular high-risk disease. Our study supports using this model for development and validation of treatments targeting PCa, especially novel immune therapeutic agents.
Read the publication to find out how QuantSeq was used to investigate radiotherapy response in prostate cancer.
Abstract
There is no strong and reliable predictive biomarker in head and neck squamous cell carcinoma (HNSCC) for EGFR inhibitors. We aimed to identify predictive and pharmacodynamic biomarkers of efficacy of afatinib, a pan-HER tyrosine kinase inhibitor, in a window-of-opportunity trial (NCT01415674). Multi-omics analyses were carried out on pre-treatment biopsy and surgical specimen for biological assessment of afatinib activity. Sixty-one treatment-naïve and operable HNSCC patients were randomised to afatinib 40 mg/day for 21–28 days versus no treatment. Afatinib produced a high rate of metabolic response. Responders had a higher expression of pERK1/2 (P = 0.02) and lower expressions of pHER4 (P = 0.03) and pRB1 (P = 0.002) in pre-treatment biopsy compared to non-responders. At the cellular level, responders displayed an enrichment of tumor-infiltrating B cells under afatinib (P = 0.02). At the molecular level, NF-kappa B signaling was over-represented among upregulated genes in non-responders (P < 0.001; FDR = 0.01). Although exploratory, phosphoproteomics-based biomarkers deserve further investigations as predictors of afatinib efficacy.
Read the publication for the full story and find out how High-throughput 3′ Tag RNA-seq tumor RNA was carried out with QuantSeq to reveal expression signatures in head and neck squamous cell carcinoma.
In addition to various other publications, our customers also shared their data several webinars which are available to you on-demand.
Webinars where our expert customers show their data on how QuantSeq was successfully used in global gene expression studies of biobanked FFPE samples
Whole Transcriptome Sequencing on FFPE Samples with CORALL: Showcases from Cancer Research
Publications using CORALL for WTS of FFPE Samples
Abstract
Atypical teratoid/rhabdoid tumors (AT/RT) are the most common malignant brain tumors manifesting in infancy. They split into four molecular types. The major three (AT/RT-SHH, AT/RT-TYR, and AT/RT-MYC) all carry mutations in SMARCB1, the fourth quantitatively smaller type is characterized by SMARCA4 mutations (AT/RT-SMARCA4). Molecular characteristics of disease recurrence or metastatic spread, which go along with a particularly dismal outcome, are currently unclear. Here, we investigated tumor tissue from 26 patients affected by AT/RT to identify signatures of recurrences in comparison with matched primary tumor samples. Microscopically, AT/RT recurrences demonstrated a loss of architecture and significantly enhanced mitotic activity as compared to their related primary tumors. Based on DNA methylation profiling, primary tumor and related recurrence were grossly similar, but three out of 26 tumors belonged to a different molecular type or subtype after second surgery compared to related primary lesions. Copy number variations (CNVs) differed in six cases, showing novel gains on chromosome 1q or losses of chromosome 10 in recurrences as the most frequent alterations. To consolidate these observations, our cohort was combined with a data set of unmatched primary and recurrent AT/RT, which demonstrated chromosome 1q gain and 10 loss in 18% (n = 7) and 11% (n = 4) of the recurrences (n = 38) as compared to 7% (n = 3) and 0% (n = 0) in the primary tumors (n = 44), respectively. Similar to the observations made by DNA methylation profiling, RNA sequencing of our cohort revealed AT/RT primary tumors and matched recurrences clustering closely together. However, a number of genes showed significantly altered expression in AT/RT-SHH recurrences. Many of them are known tumor driving growth factors, involved in embryonal development and tumorigenesis, or are cell-cycle-associated. Overall, our work identifies subtle molecular changes that occur in the course of the disease and that may help define novel therapeutic targets for AT/RT recurrences.
Read more to find out how CORALL RNA-Seq was employed to assess expression profiles in atypical teratoid/rhabdoid (AT/RT) malignant brain tumors.
Abstract
Group 3 medulloblastoma is one of the most aggressive types of childhood brain tumors. Roughly 30% of cases carry genetic alterations in MYC, SMARCA4, or both genes combined. While overexpression of MYC has previously been shown to drive medulloblastoma formation in mice, the functional significance of SMARCA4 mutations and their suitability as a therapeutic target remain largely unclear. To address this issue, we combined overexpression of MYC with a loss of SMARCA4 in granule cell precursors. Both alterations did not increase proliferation of granule cell precursors in vitro. However, combined MYC overexpression and SMARCA4 loss successfully induced tumor formation in vivo after orthotopic transplantation in recipient mice. Resulting tumors displayed anaplastic histology and exclusively consisted of SMARCA4-negative cells although a mixture of recombined and non-recombined cells was injected. These observations provide first evidence for a tumor-promoting role of a SMARCA4 deficiency in the development of medulloblastoma. In comparing the transcriptome of tumors to the cells of origin and an established Sonic Hedgehog medulloblastoma model, we gathered first hints on deregulated gene expression that could be specifically involved in SMARCA4/MYC driven tumorigenesis. Finally, an integration of RNA sequencing and DNA methylation data of murine tumors with human samples revealed a high resemblance to human Group 3 medulloblastoma on the molecular level. Altogether, the development of SMARCA4-deficient medulloblastomas in mice paves the way to deciphering the role of frequently occurring SMARCA4 alterations in Group 3 medulloblastoma with the perspective to explore targeted therapeutic options.
Read the publication for the full story and how CORALL was used for RNA-Seq in mouse models of medulloblastoma.
Abstract
Background
Myxopapillary ependymoma (MPE) is a heterogeneous disease regarding histopathology and outcome. The underlying molecular biology is poorly understood, and markers that reliably predict the patients’ clinical course are unknown.
Methods
We assembled a cohort of 185 tumors classified as MPE based on DNA methylation. Methylation patterns, copy number profiles, and MGMT promoter methylation were analyzed for all tumors, 106 tumors were evaluated histomorphologically, and RNA sequencing was performed for 37 cases. Based on methylation profiling, we defined two subtypes MPE-A and MPE-B, and explored associations with epidemiological, clinical, pathological, and molecular characteristics of these tumors.
Results
MPE-A occurred at a median age of 27 years and were enriched with tumors demonstrating papillary morphology and MGMT promoter hypermethylation. Half of these tumors could not be totally resected, and 85% relapsed within 10 years. Copy number alterations were more common in MPE-A. RNA sequencing revealed an enrichment for extracellular matrix and immune system-related signatures in MPE-A. MPE-B occurred at a median age of 45 years and included many tumors with a histological diagnosis of WHO grade II and tanycytic morphology. Patients within this subtype had a significantly better outcome with a relapse rate of 33% in 10 years (P = 3.4e-06).
Conclusions
We unraveled the morphological and clinical heterogeneity of MPE by identifying two molecularly distinct subtypes. These subtypes significantly differed in progression-free survival and will likely need different protocols for surveillance and treatment.
Dive into the publication to find out how RNA-Seq with CORALL and methylation profiling helped define two distinct subtypes of myxopapillary ependymoma (MPE) with disparate clinical behavior.
FFPE RNA EXPERTISE blog series: What is coming next?
More is yet to come, including practical tips and actionable advice on best practices for FFPE RNA-Seq experiments and a nifty checklist for a quick overview to keep at your (bench) side. Stay tuned.
References
Bessa, C.; Matos, P.; Jordan, P.; Gonçalves, V. Alternative Splicing: Expanding the Landscape of Cancer Biomarkers and Therapeutics. Int. J. Mol. Sci. 2020, 21, 9032. doi: 10.3390/ijms21239032
Bockmayr, M., Harnisch, K., Pohl, L. C., Schweizer, L., Mohme, T., Körner, M., Alawi, M., Suwala, A. K., Dorostkar, M. M., Monoranu, C. M., Hasselblatt, M., Wefers, A.K., Capper, D., Hench, J., Frank, S., Richardson, T. E., Tran, I., Liu, E., Snuderl, M., Engertsberger, L., Benesch, M., von Deimling, A., Obrecht, D., Mynarek, M., Rutkowski, S., Glatzel, M., Neumann, J. E., Schüller, S., Comprehensive profiling of myxopapillary ependymomas identifies a distinct molecular subtype with relapsing disease, Neuro-Oncology, 2022, 24, Pages 1689–1699, doi: 10.1093/neuonc/noac088
Dorney, R.; Dhungel, B. P.; Rasko, J. E. J.; Hebbard, L.; Schmitz, U. Recent advances in cancer fusion transcript detection, Briefings in Bioinformatics 2023, 24, bbac519, doi: 10.1093/bib/bbac519
French, J. D.; Edwards, S. L. The Role of Noncoding Variants in Heritable Disease. Trends in Genetics. 2020, 36, 880-891. doi: 10.1016/j.tig.2020.07.004
Göbel, C., Godbole, S., Schoof, M. et al. MYC overexpression and SMARCA4 loss cooperate to drive medulloblastoma formation in mice. acta neuropathol commun 2023, 11, 174. doi: 10.1186/s40478-023-01654-2
Haughey, C.M.; Mukherjee, D.; Steele, R.E.; Popple, A.; Dura-Perez, L.; Pickard, A.; Patel, M.; Jain, S.; Mullan, P.B.; Williams, R.; et al. Investigating Radiotherapy Response in a Novel Syngeneic Model of Prostate Cancer. Cancers 2020, 12, 2804. doi: 10.3390/cancers12102804
Johann P.D., Altendorf L., Efremova E.M., Holsten T., Steinbügl M., Nemes K., Eckhardt A., Kresbach C., Bockmayr M., Koch A., Haberler C., Antonelli M., DeSisto J., Schuhmann M.U., Hauser P., Siebert R., Bens S., Kool M., Green A.L., Hasselblatt M., Frühwald M.C., Schüller U. Recurrent atypical teratoid/rhabdoid tumors (AT/RT) reveal discrete features of progression on histology, epigenetics, copy number profiling, and transcriptomics. Acta Neuropathol. 2023, 146(3):527-541. doi: 10.1007/s00401-023-02608-7.
Marret, G., Temam, S., Kamal, M. et al. Randomized phase II study of preoperative afatinib in untreated head and neck cancers: predictive and pharmacodynamic biomarkers of activity. Sci Rep 13, 22524 (2023). doi: 10.1038/s41598-023-49887-4
Mehine, M., Ahvenainen, T., Khamaiseh, S., Härkönen, J., Reinikka, S., Heikkinen, T., Äyräväinen, A., Pakarinen, P., Härkki, P., Pasanen., A., Levonen., A., Bützow., R., Vahteristo, P. A novel uterine leiomyoma subtype exhibits NRF2 activation and mutations in genes associated with neddylation of the Cullin 3-RING E3 ligase. Oncogenesis 2022, 11, 52. doi: 10.1038/s41389-022-00425-3
Taber A., Christensen E., Lamy P., Nordentoft I., Prip F., Lindskrog SV., Birkenkamp-Demtröder K., Okholm T.L.H., Knudsen M., Pedersen J.S., Steiniche T., Agerbæk M., Jensen J.B., Dyrskjøt L. Molecular correlates of cisplatin-based chemotherapy response in muscle invasive bladder cancer by integrated multi-omics analysis. Nat Commun. 2020 11(1):4858. doi: 10.1038/s41467-020-18640-0. Erratum in: Nat Commun. 2022 Apr 4;13(1):1916. doi: 10.1038/s41467-022-29627-4. PMID: 32978382; PMCID: PMC7519650.
Turnbull, A.K., Selli, C., Martinez-Perez, C. et al. Unlocking the transcriptomic potential of formalin-fixed paraffin embedded clinical tissues: comparison of gene expression profiling approaches. BMC Bioinformatics 2020, 21, 30. doi: 10.1186/s12859-020-3365-5
Quintela-Fandino M., Holgado E., Manso L., Morales S., Bermejo B., Colomer R., Apala J.V., Blanco R., Muñoz M., Caleiras E., Iranzo V., Martinez M., Dominguez O., Hornedo J., Gonzalez-Cortijo L., Cortes J., Gasol Cudos A., Malon D., Lopez-Alonso A., Moreno-Ortíz M.C., Mouron S., Mañes S.. Immuno-priming durvalumab with bevacizumab in HER2-negative advanced breast cancer: a pilot clinical trial. Breast Cancer Res. 2020, 22:124. doi: 10.1186/s13058-020-01362-y. PMID: 33176887; PMCID: PMC7661209.
Written by Masa Ivin, PhD and Dr. Yvonne Goepel
Photographs courtesy of Mag. Amra Dedic