Human-derived specimens obtained during a biopsy are a potent source of disease-relevant molecular information. The information stored in these specimens is essential for diagnosing and choosing treatments and fuelling innovative biomedical research. The advancement of personalized cancer treatment, innovative therapies, biomarker discovery and research, and target identification is currently primarily reliant on the availability of these biopsy-derived human tissue samples.
Following biopsy analysis, preserved tissue samples are collected in hospital-associated biobanks, research centers, or private companies supporting biomedical research. These clinical biopsy tissue samples come in many forms, and some are more suitable for next-generation sequencing (NGS) studies than others.
Formalin-fixed paraffin-embedded (FFPE) and fresh frozen (FF) are the most common methods to preserve human solid tissues. The FFPE approach uses formalin to halt cellular processes and “fix” tissues, whereas FF sample preparation relies on rapid cooling-down to achieve the same goal. Both approaches have their own advantages and disadvantages, and choosing one will depend on the study’s requirements, research questions, methodology and logistic challenges associated to the project.
While frozen samples are easier to work with when it comes to NGS approaches (both DNA- and RNA-Seq), FFPE samples have other advantages that make them more suitable despite being more challenging to work with (Sah et al., 2013). The good news is that new technologies and FFPE-specific protocols can help overcome many of these challenges, enabling reliable NGS data from FFPE samples.
We frequently receive inquiries about the benefits and drawbacks of utilizing FFPE and FF samples for molecular analysis, especially RNA-Seq, so we decided to dive deeper into the matter in this blog article.
Fresh frozen samples
Cryopreservation is a process that preserves organelles, cells, tissues, or any other biological constructs by cooling the samples to very low temperatures. To induce immediate cryopreservation, biopsy tissue specimens are immersed in liquid nitrogen and stored in ultra-cold freezers at -80°C. This process is known as “flash-freezing” or “snap-freezing”. If prepared and stored correctly, nucleic acids in fresh frozen samples are perfectly preserved, ideal for downstream next-generation sequencing analysis.
The quality of DNA and RNA isolated from FF tissues is higher than FFPE-derived DNA and RNA, making fresh frozen samples a gold standard for NGS. As well, compared to FFPE samples, FF samples do not require laborious preparation, as the process of freezing tissue is less complicated, less time-consuming, and does not involve toxic chemicals such as formalin. However, there are a few practical hurdles when it comes to preparing and storing frozen tissue samples, and we will point out some of them. These hurdles are the reason why fresh frozen sample biobanks are not widely available (Jacobsen et al., 2023). Because of their low availability, they are rarely used for large-scale retrospective studies of tumors — studies that will ultimately drive the development of new cancer therapies.
Disadvantages of fresh frozen samples
- Sampling issues
To promptly freeze biopsy tissues during surgeries, it is necessary to have liquid nitrogen containers and a -80°C freezer close to surgery rooms. In practice, it is rarely possible to provide such conditions. - Complicated and costly storage
Tissue degrades quickly at room temperature and does not store well when kept in more standard -20 °C food or household freezers. A dedicated ultra-low-temperature freezer (-80°C) is needed to keep the tissue frozen. In addition, archives of frozen tissues are vulnerable to power outages or mechanical failures. Frozen collections cannot last as long as FFPE ones; maintenance is way more costly and demanding. - Mistakes due to a human factor
If samples are left on laboratory benches for too long or freezer doors are not properly closed, these precious frozen tissue collections could be destroyed.
Comparison between FFPE vs Fresh Frozen Samples
In our blog “What are FFPE samples and how are they prepared?” we have provided a comprehensive overview of FFPE samples, their preparation, and the challenges they pose in various downstream applications. In the article “Applications of FFPE samples in modern research” we looked into the specific challenges encountered when using FFPE samples in NGS studies, a topic of significant interest and importance in our field.
Here, we will discuss the pros and cons of next-generation sequencing applications by comparing them to fresh frozen samples.
The most significant advantage of FFPE samples over FF samples is their wide availability. FFPE samples are routinely collected and archived during patient treatment and care for many decades now, and there are 400 million (Sah et al., 2013) to probably more than a billion (Blow, 2007) FFPE samples in hospitals and biobanks worldwide. If one looks at solid tumor biobanks, the estimates are that between 50 and 80 million FFPE samples are suitable for NGS. Many are clinically described— primary diagnosis, therapeutic regimen, drug response, and recurrence status are known— so they can be linked to clinical outcomes and long-term follow-up. Despite this enormous clinical resource, much of it has long remained unexplored by NGS – because FFPE samples usually harbor only small amounts of low-quality nucleic acids. Indeed, the biggest bottleneck to high-quality NGS from FFPE samples is obtaining high-quality DNA and RNA. Chemical modifications, fragmentation of DNA and RNA, and cross-linking to proteins introduced through the fixation process can all negatively impact the data quality. This is why solutions and workflows specifically optimized for FFPE samples are game changers when it comes to the quality of FFPE DNA-Seq or RNA-Seq data.
Is NGS data quality comparable between FFPE and FF samples?
According to the screened literature (and our extensive experiments – see below), NGS data quality obtained with FFPE samples matches data quality of NGS with fresh frozen samples (Hedegaard et al., 2014; van Allen et al., 2014).
Many studies have compared FFPE and fresh frozen samples regarding different types of DNA-Seq. Whole exome sequencing (WES) is a powerful method that can positively impact the care of cancer patients if properly implemented into clinics (van Allen et al., 2014; Astolfi et al., 2015). A study published in Nature Medicine aimed to compare the quality of WES data from FFPE vs. frozen samples taken from identical lung adenocarcinoma tumors (van Allen et al., 2014). They could demonstrate robust WES using FFPE-derived tumor DNA with detected alterations comparable to those detected in fresh samples.
Whole genome sequencing (WGS) is a comprehensive method for genome analysis, capable of identifying novel genome events without prior knowledge of targets. Fresh frozen tissue is the optimal source of DNA for WGS of cancer patients. While fresh frozen tissue remains the gold standard for WGS in cancer patients, a prospective study for the 100,000 Genomes Project explored the use of FFPE samples as a DNA source (Robbe et al., 2018). Despite the identified shortcomings of FFPE samples compared to fresh frozen, these issues can be mitigated through optimized DNA extraction and bioinformatics analysis, supporting the use of FFPE cancer samples in clinical settings.
Just as with DNA-Seq, studies comparing the RNA-Seq data quality of FFPE and fresh frozen tissue samples are of great importance. These studies play a crucial role in determining the reliability of FFPE transcriptional profiles for cancer research studies, among others. In one of the studies, by optimizing a pipeline specifically for FFPE RNA-Seq (Pennock et al., 2019), authors could use FFPE breast cancer tissues to distinguish between estrogen receptor positive (ER+) and estrogen receptor negative (ER-) breast cancer cases. They confirmed gene expression data obtained from FFPE samples to publicly available databases obtained from fresh tissues.
In 2020, we have hosted a webinar in which Dr. Andrew Beggs (University of Birmingham, UK) gave a talk on the importance of RNA-Seq in the multi-omics approach in clinical cancer diagnostics. He introduced his lab’s project, where they looked at combination of whole transcriptome sequencing with 3′ mRNA-Seq as an approach to get interesting insights into the underlying disease processes in colorectal cancer. His team performed whole-genome sequencing on fresh cancer tissue samples and, as a proof of principle, 3’ mRNA-Seq (using our QuantSeq) on matched formalin-fixed, paraffin-embedded samples.
Experiments from Lexogen’s bench: Comparison between RNA-Seq from FFPE vs fresh frozen tissues
While adapting our RNA-Seq workflows to FFPE samples and developing our FFPE Transcriptomics line we ran many FFPE RNA-Seq experiments in our R&D, including several “comparison” experiments to ensure transcriptome profiles obtained from FFPE samples match those of fresh frozen samples.
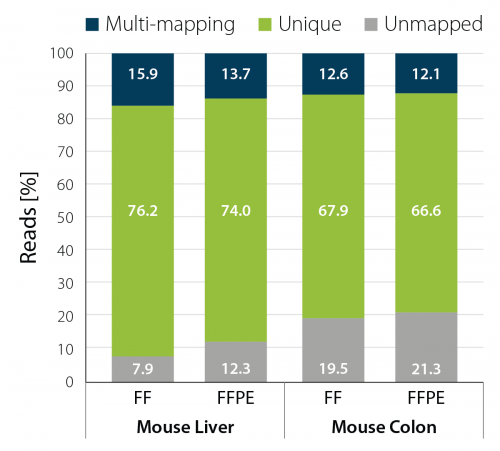
Among those is an experiment where we looked at the mapping statistics for libraries generated from different mouse tissues comparing FFPE and FF sample tissues. As shown in the Fig. 1, in both mouse liver and mouse colon, the percentage of uniquely mapped reads is comparable between libraries obtained from FF and FFPE samples.
We are always interested in the gene detection to ensure our technologies and kits provide a sensitive assay to detect genes of all abundance classes. Using our CORALL FFPE kit for whole transcriptome sequencing from FFPE samples, we have generated libraries from mouse liver and colon, both coming from FFPE and FF samples. We consistently observed a significant overlap in the numbers of detected genes between FFPE and FF samples. A representative example is shown in Fig. 2.
Figure 2 | Overlaps of detected protein-coding genes between fresh frozen (FF) and FFPE tissue for mouse liver and colon. FFPE tissue was extracted with SPLIT One-step FFPE RNA extraction. Matched fresh reference tissue was stabilized in RNA/DNA Defender and extracted with SPLIT RNA extraction kit. Libraries were prepared using CORALL FFPE and RiboCop for Human/Mouse/Rat (HMR) V2. Samples were sequenced in paired-end mode on the Aviti Sequencer (2×76 bp). A total of 4.5 M reads was analyzed per sample. Genes detected in each of the two replicates were considered in the comparison fresh vs. FFPE. For gene detection, a CPM threshold corresponding to ≥10 reads per gene was used (i.e., at least 10 reads are required for a gene to be considered as expressed).
FFPE RNA EXPERTISE blog series: What is coming next?
In our next blog article, we will focus on different methods for RNA-Seq comparing whole transcriptome and 3’ mRNA-Seq and provide examples for their use and how to decide which method is ideal for your project.
References
Astolfi, A., Urbini, M., and Indio, V.; Nannini, Margherita; Genovese, Chiara Giusy; Santini, Donatella; Saponara, Maristella; Mandrioli, Anna; Ercolani, Giorgio; Brandi, Giovanni, et al.201516 (2015). Whole exome sequencing (WES) on formalin-fixed, paraffin-embedded (FFPE) tumor tissue in gastrointestinal stromal tumors (GIST). BMC Genomics 16, 892. doi: 10.1186/s12864-015-1982-6.
Blow, N.2007448 (2007). Tissue preparation: Tissue issues. Nature 448, 959-963. doi: 10.1038/448959a.
Hedegaard, J., Thorsen, K., and Lund, M.K.; Hein, Anne-Mette K.; Hamilton-Dutoit, Stephen Jacques; Vang, Søren; Nordentoft, Iver; Birkenkamp-Demtröder, Karin; Kruhøffer, Mogens; Hager, Henrik, et al.20149 (2014). Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS One 9, e98187. doi: 10.1371/journal.pone.0098187.
Jacobsen, S.B., Tfelt-Hansen, J., and Smerup, M.H.; Andersen, Jeppe Dyrberg; Morling, Niels202318 (2023). Comparison of whole transcriptome sequencing of fresh, frozen, and formalin-fixed, paraffin-embedded cardiac tissue. PLoS One 18, e0283159. doi: 10.1371/journal.pone.0283159.
Pennock, N.D., Jindal, S., and Horton, W.; Sun, Duanchen; Narasimhan, Jayasri; Carbone, Lucia; Fei, Suzanne S.; Searles, Robert; Harrington, Christina A.; Burchard, Julja, et al.201912 (2019). RNA-seq from archival FFPE breast cancer samples: molecular pathway fidelity and novel discovery. BMC Medical Genomics 12, 195. doi: 10.1186/s12920-019-0643-z.
Robbe, P., Popitsch, N., and Knight, S.J.L.; Antoniou, Pavlos; Becq, Jennifer; He, Miao; Kanapin, Alexander; Samsonova, Anastasia; Vavoulis, Dimitrios V.; Ross, Mark T., et al.201820 (2018). Clinical whole-genome sequencing from routine formalin-fixed, paraffin-embedded specimens: pilot study for the 100,000 Genomes Project. Genetics in Medicine 20, 1196-1205. doi: 10.1038/gim.2017.241.
Sah, S., Chen, L., and Houghton, J.; Kemppainen, Jon; Marko, Adam C.; Zeigler, Robert; Latham, Gary J.20135 (2013). Functional DNA quantification guides accurate next-generation sequencing mutation detection in formalin-fixed, paraffin-embedded tumor biopsies. Genome Medicine 5, 77. doi: 10.1186/gm481.
van Allen, E.M., Wagle, N., and Stojanov, P.; Perrin, Danielle L.; Cibulskis, Kristian; Marlow, Sara; Jane-Valbuena, Judit; Friedrich, Dennis C.; Kryukov, Gregory; Carter, Scott L., et al.201420 (2014). Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nature Medicine 20, 682-688. doi: 10.1038/nm.3559.
Written by Masa Ivin, PhD