Just Keep Swimming –
and effectively deplete rRNA with RiboCop!
RiboCop for Fish
Total RNA from fish species is comprised of large amounts of undesired ribosomal RNA (rRNA) which accounts for ~86 % of all transcripts. Lexogen’s RiboCop rRNA Depletion Kits for Fish remove undesired cytoplasmic (5.8S, 18S and 28S) and mitochondrial (5S, mt12S and mt16S) rRNA from zebrafish Danio rerio and other fish species. Ribo-depletion affords an unbiased view of the transcriptome, including non-coding and non-polyadenylated RNA transcripts.
RiboCop for Fish is ideally suited for Next Generation Sequencing (NGS) approaches in combination with CORALL RNA-Seq V2 Library Preparation Kits. RiboCop Kits are also compatible with other random primed total RNA library prep kits RNA library prep kits and long-read sequencing protocols.
Performance
Sequence what matters most and explore coding and non-coding transcripts! RiboCop enables highly effective, non-enzymatic depletion of ribosomal RNA and preserves RNA integrity for compatibility with long-read sequencing workflows.
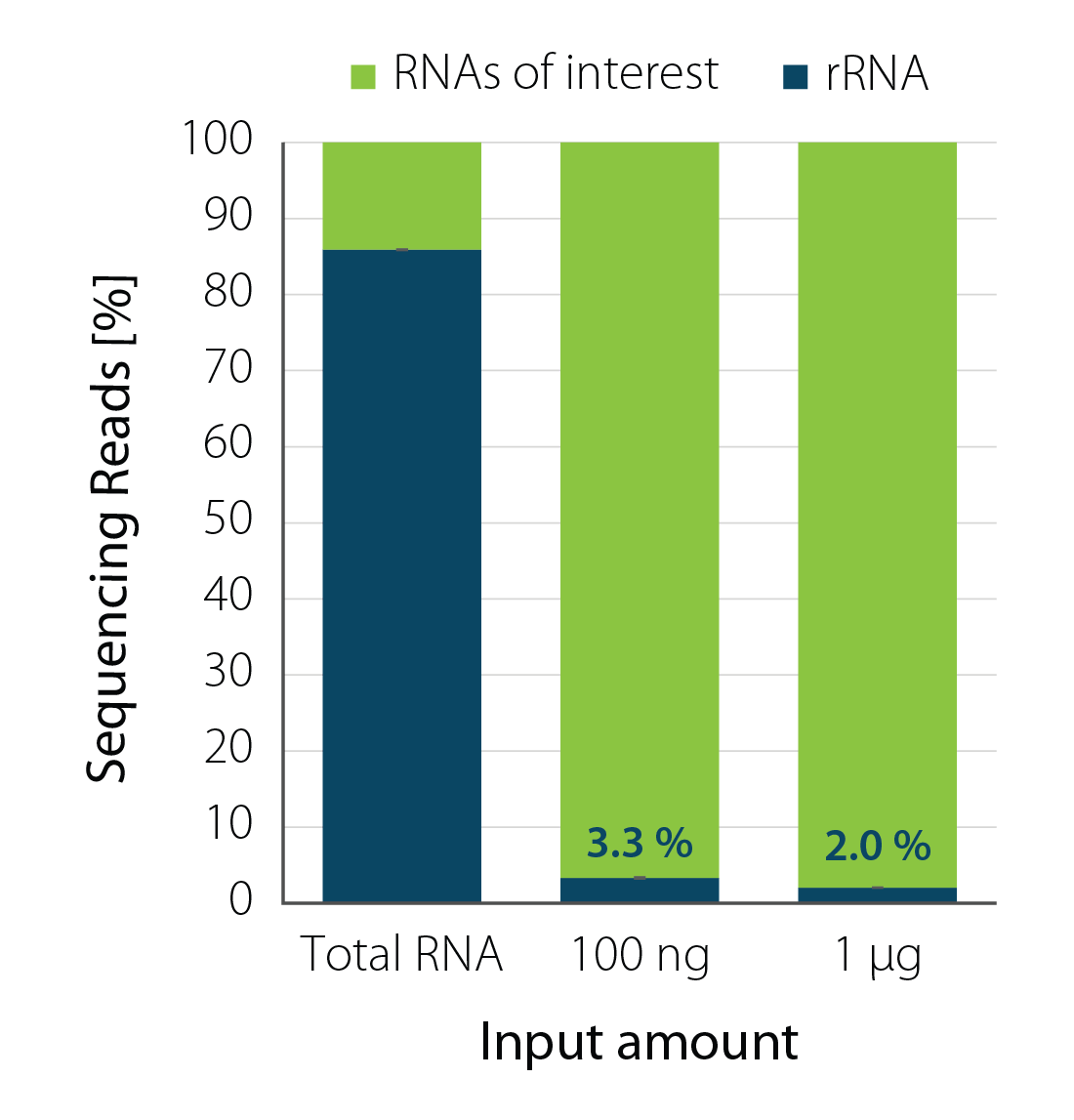
Efficient Depletion across RNA Input Amounts
Ribosomal RNA was depleted from Danio rerio total RNA using RiboCop for Fish with 100 ng or 1 µg RNA input amount and compared to total untreated RNA. RiboCop for Fish efficiently reduced rRNA reads for all input amounts (Fig. 1). RiboCop for Fish was also successfully used on further fish species, such as Gasterosteus aculeatus and Perca fluviatilis (reduction to <2 % rRNA reads for 100 ng input RNA).
rRNA Depletion Frees up Sequencing Space and Increases Unique Reads
Depletion of Danio rerio rRNA with RiboCop for Fish significantly increases uniquely mapping sequencing reads (Fig. 2). Concomitantly, the percentage of reads mapping to protein-coding genes is elevated (Table 1) delivering useful information for further analysis.
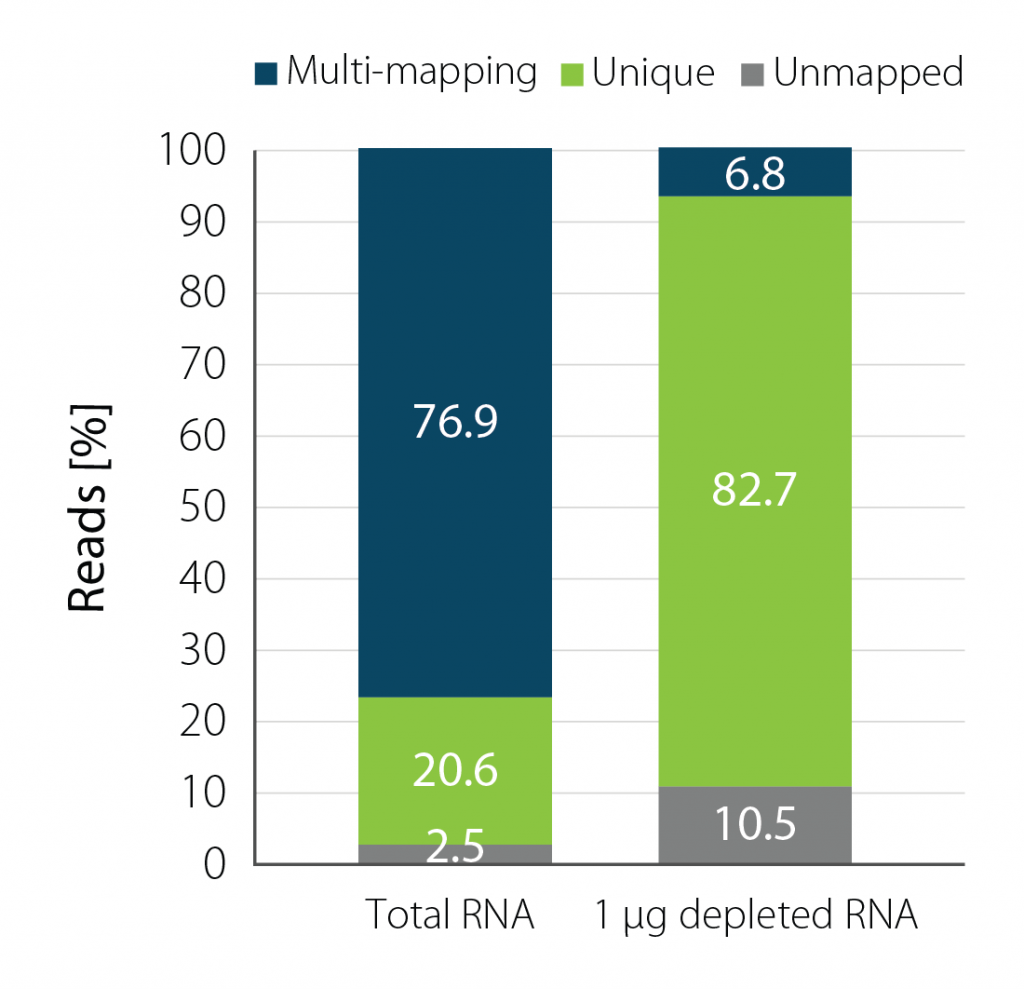
Figure 2 | Basic Mapping Stats for Total and Depleted Zebrafish RNA Input. 1 µg D. rerio RNA was depleted with RiboCop for Fish and CORALL libraries were generated from the equivalent of 10 ng total RNA using either untreated or rRNA-depleted input RNA. Libraries were sequenced on NextSeq2000 (1×90 bp) and read statistics were compared between untreated total RNA and depleted D. rerio RNA. Upon depletion, the percentage of unique reads increases 4-fold.
Table 1 | Basic Mapping Stats for RNA-Seq Libraries. Data generated from untreated zebrafish RNA vs. rRNA-depleted input RNA after RiboCop for Fish. Danio rerio RNA was processed as described in Fig. 2.
| RiboCop for Fish and CORALL RNA-Seq | ||
|---|---|---|
| D. rerio | untreated total RNA | RiboCop-depleted RNA |
| Mapped Reads | 97.5% | 89.5% |
| Multimapping Reads | 76.9% | 6.8% |
| Reads Mapped to rRNA | 85.9% | 2.0% |
| Uniquely Mapped Reads | 20.6% | 82.7% |
| Percentage of Unique Reads Mapped to Protein-coding Genes | 51.4% | 95.0% |
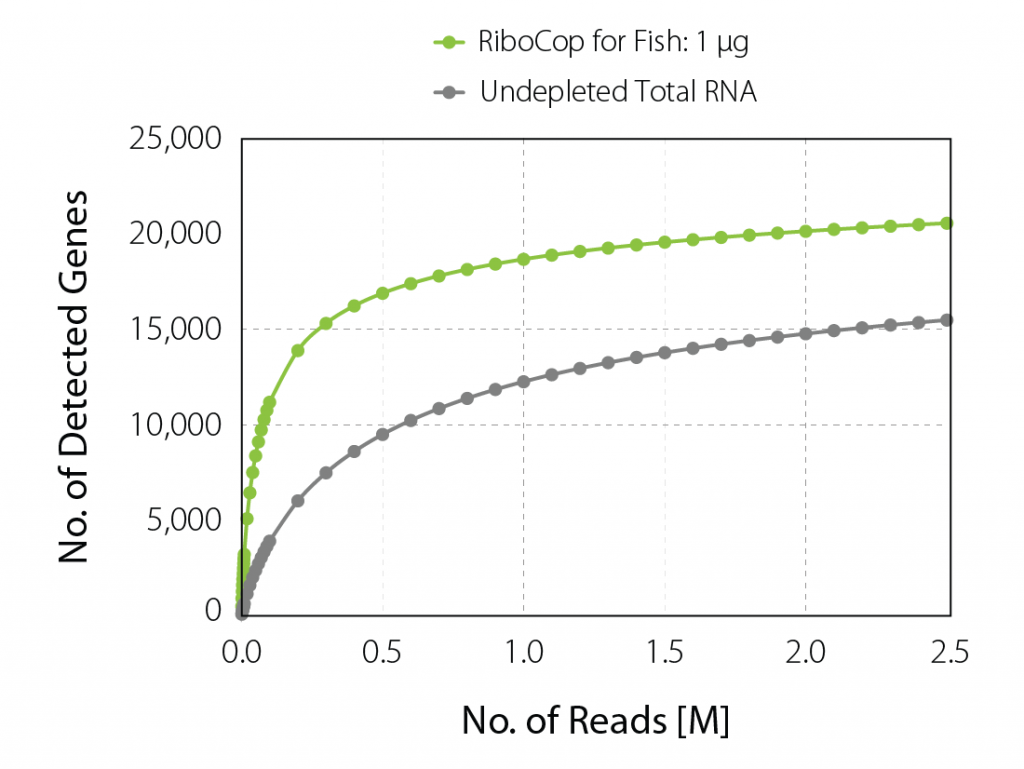
Increased Gene Detection upon Ribodepletion
Sequence what matters to your experiment. RiboCop for Fish affords an unobstructed view of the fish transcriptome and significantly increases gene discovery at lower read depth through efficient removal of ribosomal rRNA sequences (Fig. 3).
Upon rRNA depletion with RiboCop, the percentage of unique reads mapping to exons is increased 4-fold (Table 2). Concomitantly, ~14.2 K genes are detected with only 3 M reads per sample (at a threshold of CPM> 5). In contrast, only 11.9 K genes are detected without rRNA depletion at a 2.6-fold higher read depth of 7.9 M.
Table 2 | RiboCop for Fish Increases the Percentage of Unique Reads Mapping to Genes 4-fold. Significantly more genes are detected at a lower read depth for zebrafish samples upon ribo-depletion.
| RiboCop for Fish and CORALL RNA-Seq | ||
|---|---|---|
| D. rerio | untreated total RNA | RiboCop-depleted RNA |
| Sequencing Read Depth | 7.9 M | 3 M |
| No. of Detected Genes (CPM > 1) | 16,293 | 19,035 |
| No. of Detected Genes (CPM > 5) | 11,972 | 14,289 |
| Unique Reads on Genes (Exons) | 14.8% | 60.8% |
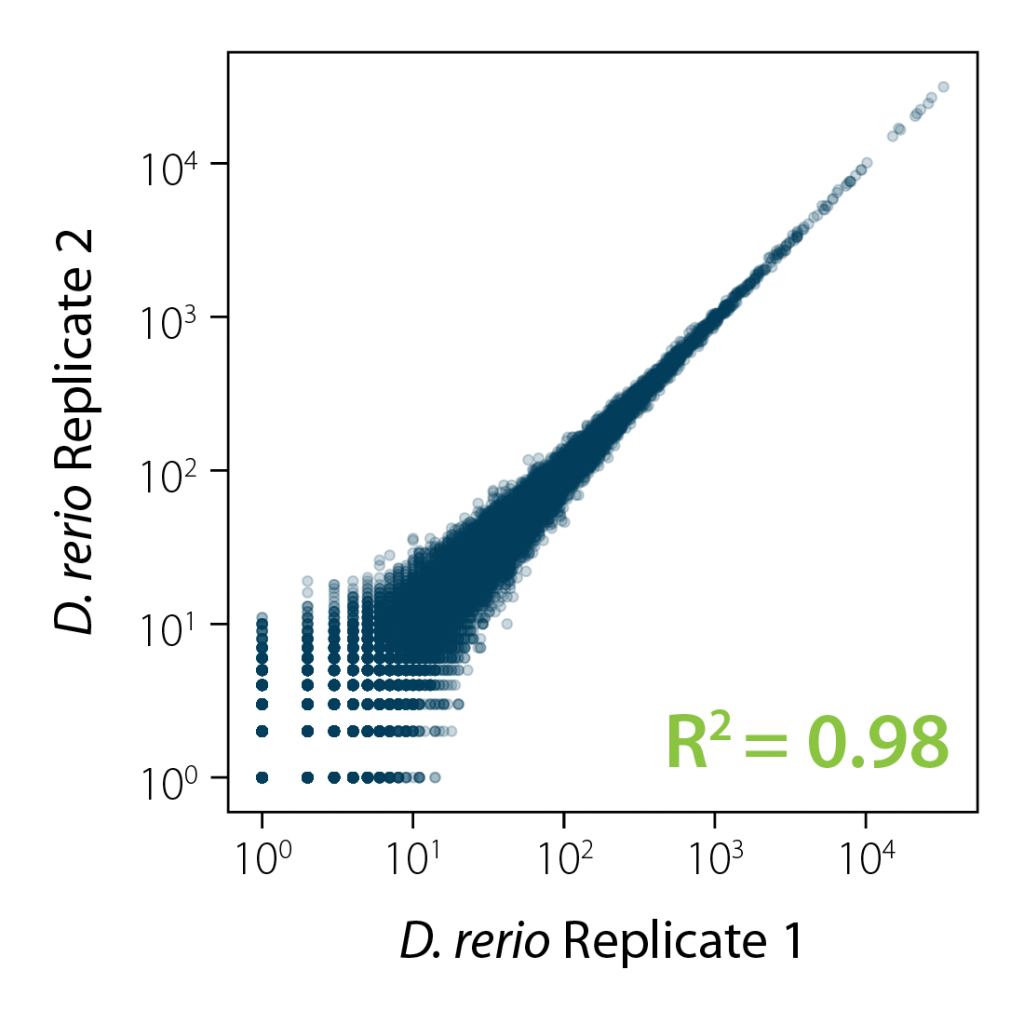
Robust Performance and High Reproducibility for Replicate Samples
RiboCop protocols are robust and highly reproducible. Correlation plots demonstrate excellent reproducibility between replicates from independent depletion reactions (Fig. 4).
Figure 4 | Excellent Reproducibility Between Replicates. Correlation of replicates for two independent depletion reactions of 1 µg D. rerio total RNA. NGS libraries were prepared using Lexogen’s CORALL RNA-Seq V2 Library Prep Kit and sequenced on NextSeq2000 (1×90 bp). Correlation analysis was performed using 3 M reads per sample.
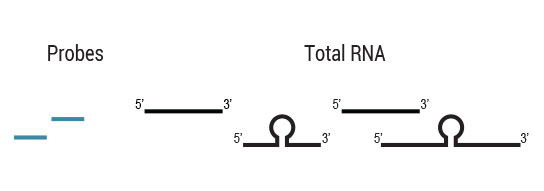
Workflow
Samples are treated using a set of affinity probes for specific depletion of rRNA sequences. RiboCop probes efficiently remove rRNA and therefore afford a comprehensive view of transcriptome composition.
1hr 30min
30 min
FAQ
Frequently Asked Questions
Access our frequently asked question (FAQ) resources via the buttons below.
Please also check our General Guidelines and FAQ resources!
How do you like the new online FAQ resource? Please share your feedback with us!
Downloads
Safety Data Sheet
If you need more information about our products, please contact us through support@lexogen.com or directly under +43 1 345 1212-41.
First time user of RiboCop?
First Time User? We’re excited to offer you an exclusive introductory offer.
Buy from our Webstore
Need a web quote?
You can generate a web quote by Register or Login to your account. In the account settings please fill in your billing and shipping address. Add products to your cart, view cart and click the “Generate Quote” button. A quote in PDF format will be generated and ready to download. You can use this PDF document to place an order by sending it directly to sales@lexogen.com.
Web quoting is not available for countries served by our distributors. Please contact your local distributor for a quote.