Do you have any questions?
Find the ideal kit for your application:
SLAMseq Catabolic Kinetic RNA-Seq
SLAMseq Catabolic RNA-Seq labels newly synthesized (nascent) transcripts in cell culture experiments and detects the decreasing signal of nascent RNA as they are being degraded.
Precise time-resolved analysis of RNA degradation and turnover
SLAMseq offers a straightforward and easy approach to track signal reduction over time for precise and time-resolved measurements of RNA decay in response to stimuli. Compared to conventional methods, SLAMseq delivers higher resolution and allows to study treatment effects and their sequence within cellular processes. SLAMseq Catabolic RNA-Seq is ideal for transcript stability and mRNA half-life measurements, to decipher cell-signaling pathways, or during drug discovery and development processes, and target identification & validation or biomarker discovery.
Performance
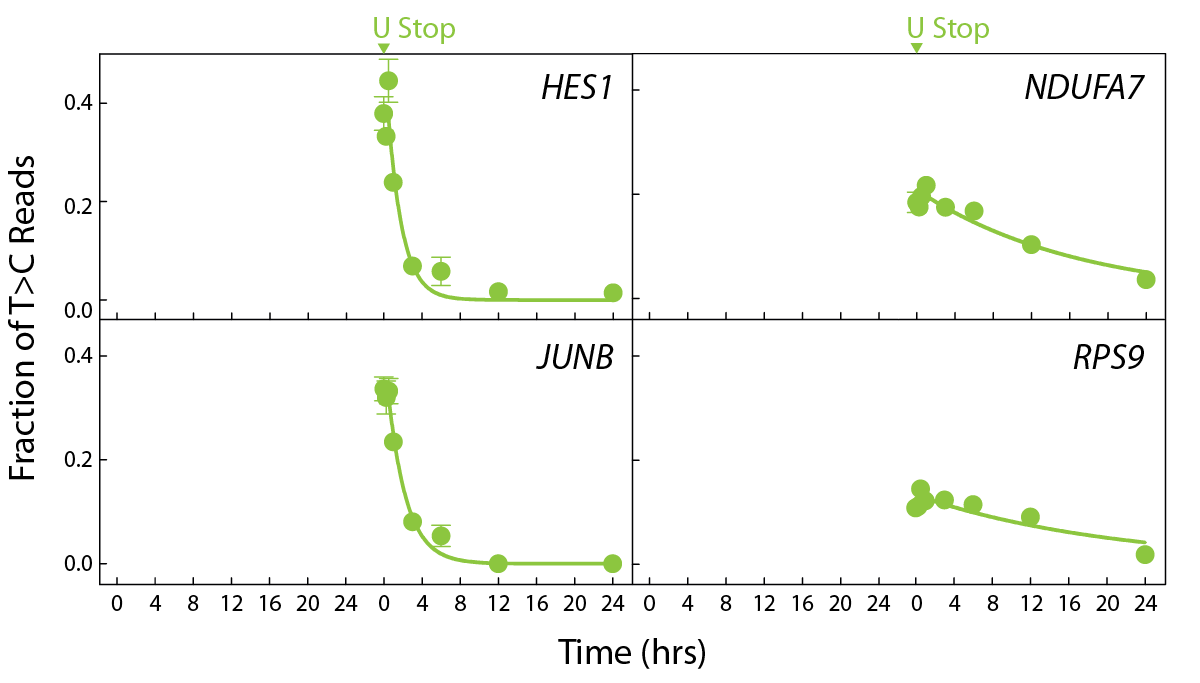
SLAMseq delivers precise measurements for global RNA decay rates in living cells and allows to distinguish primary and secondary effects of treatments.
Catabolic Kinetics Labeling of RNA
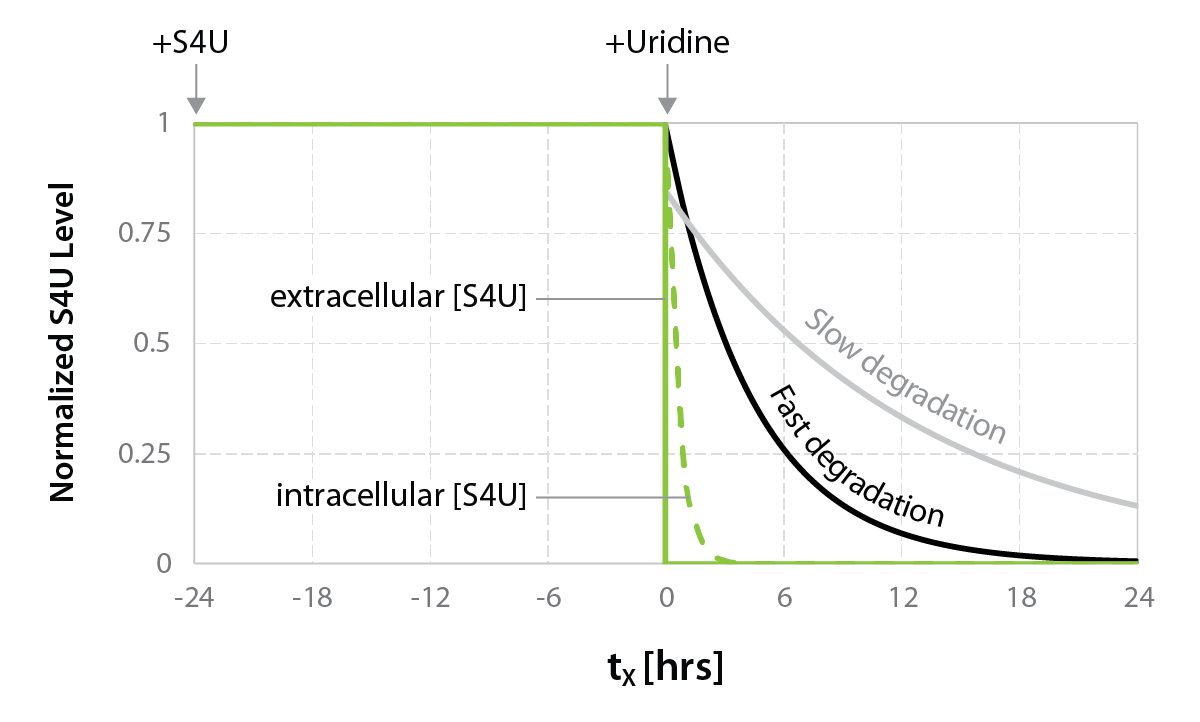
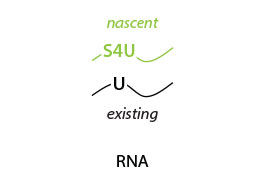
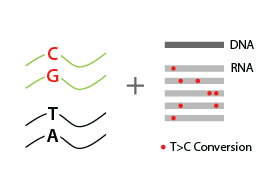
The Catabolic Kinetics Module uses a longer initial S4U labeling duration to enable RNA metabolism to reach an approximate steady-state level. The exchange of S4U for unlabeled uridine in cell media stops the labeling at t0 (chase) and samples are collected over time (tx up to 24 hours) following addition of unlabeled uridine. Existing RNA is therefore labeled with S4U, while nascent RNA synthesized after the uridine chase is unlabeled. The experiment is thus ideal to monitor RNA degradation rates (Fig. 1).
Monitor RNA Degradation Rates
The SLAMseq Kinetics Kit – Catabolic Kinetics Module is ideal to measure transcript degradation rates. Cells are first grown in S4U-containing medium for a specified time period to label existing RNA. S4U is then replaced by unmodified uridine (+Uridine) and RNA is sampled. RNA degradation rates are calculated from the decrease in S4U-labeled RNA over time (Fig. 2). For cost-efficient RNA-Seq analysis of degradation rates convenient bundles of SLAMseq Catabolic Kinetic 3’ mRNA-Seq with QuantSeq FWD V2 are available.
Analyze Transcriptome-Wide Expression Dynamics
SLAMseq resolves transcript expression dynamics on a transcriptome-wide scale. Individual transcript RNA degradation rates can be measured directly. RNA degradation rates are depending on gene regulation processes, such as blockers, enhancer, cofactors, and are transcript specific.
Identify Direct Transcriptional Targets of Any Gene
Combining SLAMseq with protein modulators, or drug treatments distinguishes direct (primary) and indirect (secondary) target responses. Use SLAMseq to dissect signaling pathways underlying biological processes and characterize drug-target responses on the transcriptional level (Muhar, M et al., 2018).
Find more information about SLAMseq data analysis here or contact us at support@lexogen.com.
Workflow
SLAMseq Catabolic Kinetic RNA-Seq allows to detect RNA degradation rates over time in cell culture experiments in response to a stimuli, e.g., a fast-acting compound or drug candidate. Optimal concentration of labeling reagent must be determined during exploring phase to avoid cell toxicity.
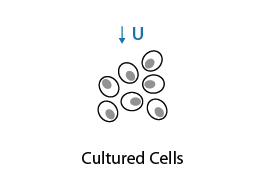
Cultured cells are grown in 4-Thiouridine (S4U) for up to 24 h to allow S4U nucleotides to be incorporated into newly synthesized RNA until saturation is reached. Optimal concentration of S4U must be determined during exploring phase to avoid cell toxicity.
Cells are treated for >30 min with fast-acting compound or drug. The incubation duration depends on the compound and cell type. Negative controls need to be applied.
Cells are supplied with fresh media containing Uridine to perform a chase experiment to assess RNA decay over time.
Cells are sampled at a given timepoint(s) and RNA is extracted under reducing conditions. The isolated total RNA contains both existing (labeled) and newly synthesized (unlabeled) RNA for catabolic kinetics experiments.
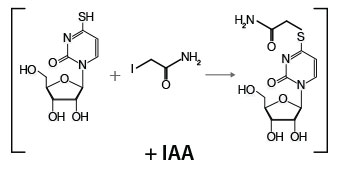
Total RNA (1 - 5 µg) is mixed with iodoacetamide (IAA), which modifies the 4-thiol group of S4U-containing nucleotides via the addition of a carboxyamidomethyl group. The RNA is purified by ethanol precipitation prior to proceeding to library preparation.
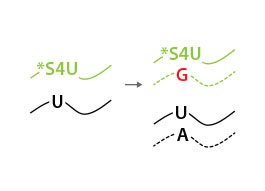
Total RNA is reverse-transcribed to cDNA. For labeled RNA transcripts, reverse transcriptase incorporates a G instead of an A at positions where reduced *S4U-modified nucleotides are encountered.
Second-strand cDNA synthesis is then performed to generate a double-stranded SLAMseq RNA-Seq library.
FAQ
Frequently Asked Questions
Access our frequently asked question (FAQ) resources via the buttons below.
Please also check our General Guidelines and FAQ resources!
How do you like the new online FAQ resource? Please share your feedback with us!
Downloads
Safety Data Sheet
If you need more information about our products, please contact us through support@lexogen.com or directly under +43 1 345 1212-41.
Ordering Information
| Cat. № | Product Name | ||
|---|---|---|---|
| 062.24 | SLAMseq Kinetics Kit - Catabolic Kinetics Module, 24 preps | SLAMseq Kinetics Kit - Catabolic Kinetics Module, 24 preps | |
| 230.24 | SLAMseq Catabolic Kinetic 3’ mRNA-Seq with QuantSeq FWD V2, 24 preps | SLAMseq Catabolic Kinetic 3’ mRNA-Seq with QuantSeq FWD V2, 24 preps | |
First time user of SLAMseq?
First Time User? We’re excited to offer you an exclusive introductory offer.

Not sure how to get started with your SLAMseq experiment?
Try the SLAMseq product configurator and find the ideal setup for your application!
Buy from our Webstore
Need a web quote?
You can generate a web quote by Register or Login to your account. In the account settings please fill in your billing and shipping address. Add products to your cart, view cart and click the “Generate Quote” button. A quote in PDF format will be generated and ready to download. You can use this PDF document to place an order by sending it directly to sales@lexogen.com.
Web quoting is not available for countries served by our distributors. Please contact your local distributor for a quote.